Publication – Improving Bacteria’s Reception of Biomolecules
Tags:
Outline the paper
Imagine a future where we could use bacteria to create sensors and computers that utilize their ability to exchange molecules. In this scenario we would be able to use these cells to monitor our environment, or even diagnose and treat some human diseases. In this short paper we contribute to this goal by investigating bacterial internal signal processing using analog electronics.
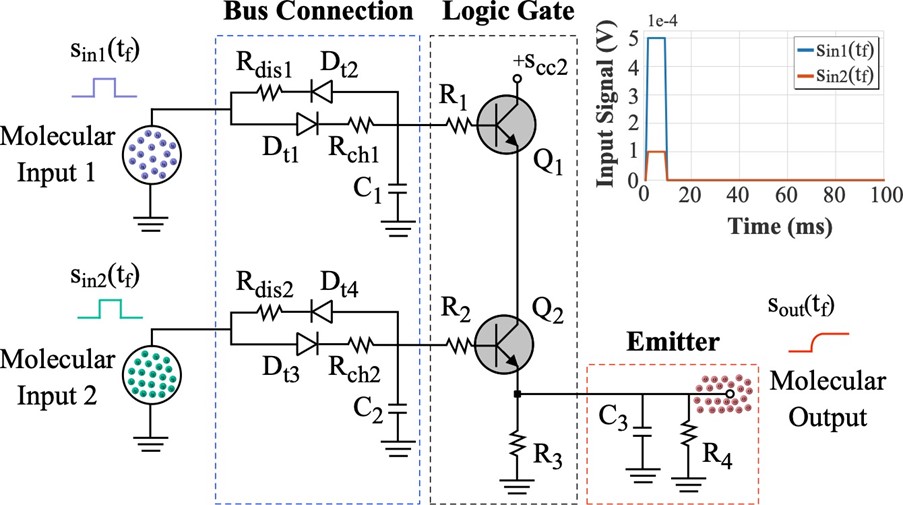
Bacteria populations consume a variety of molecules to ensure their survival, including sugars and proteins. This process is facilitated by the receptors that each bacterium cell has in their membrane, which specifically collect the molecules that are available in the environment that surrounds them. In this paper we propose the use of an electronic circuit to represent the molecular consumption by bacterial populations, which is based on the design of an analogue AND logic gate, and it is combined with two other circuits to represent the molecular binding (initial part of the circuit) and the diffusion of the secondary signal within the bacterial population.
Who will it help?
By investigating the bacteria molecular binding we are supporting the development of biosensors using bacteria with high sensitivity to detect lower concentrations of pollutants and chemicals that can harm the soil. Therefore, this research can support the design of biocompatible tools for the better management and use of natural resources, which will be fundamental for the dairy industry in the future. Moreover, the proposed circuit representation will allow biotechnologists to create rational designs of cells that will further our knowledge of those systems. Furthermore, through this approach researchers can identify limitations and opportunities for their applications before starting to setup their wet lab experiments.
What is the future of this research?
For the next step in this research, we will investigate other aspects of bacteria engineering using the analogue circuit representation, such as their internal signaling processing systems required for the execution of collective behaviour. We also plan to improve the characterisation of every aspect of the circuit to improve the accuracy of this representation, when compared to the real cell. The aim is to make a robust model that can used by other researchers to design and develop biotechnological applications without fully relying on wet lab experiments. This would become an in-silico prototyping platform to engineer bacterial populations for a wide variety of applications, ranging from simple biosensors up to complex biocomputing systems.
Currently, the proposed approach can be utilised as the first step for the design biosensors using bacterial populations, which includes the characterisation of the molecule detection and production of secondary signals. The results from this initial analysis can be further refined through wet lab experiments to develop this bacteria-based biosensors. In the future, together with the evolution of the synthetic biology field, the circuit representation of bacterial populations will be fundamental for the development of specialised and improved bioreactors for drug design, intrabody health monitors for animal and human’s digestive system, and biocomputing systems for biological data processing and storage.
Publication Title: Binding Process Analysis of Bacterial-based AND Logic Gates
Authors: Daniel Perez Martins, Paul D. Cotter, Orla O’Sullivan and Sasitharan Balasubramaniam
Publication Date: 07 Sept 2021
Journal: Eight Annual ACM International Conference on Nanoscale Computing and Communication
Link to publication: https://dl.acm.org/doi/10.1145/3477206.3477472




