Publication – Creating Binary Codes Using Bacteria
Tags:
Outline the paper
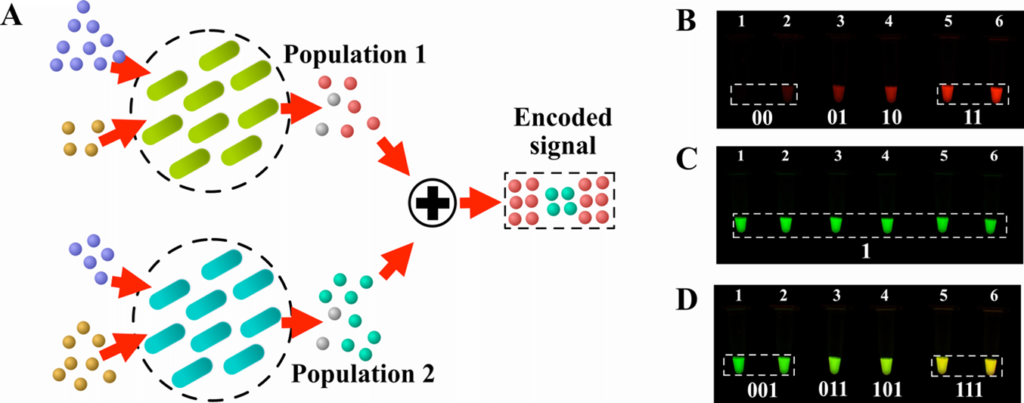
Modern digital systems, like CDs and computers, utilise codes to transform the binary data they store or transmit to protect them against errors produced by the environment where the data is stored/propagated. We envisioned a similar system to protect the signals (i.e. molecules) produced by bacterial populations in the environments that they are found. The proposed model considers two bacterial populations, one emitting molecules and the other encoding them (i.e. creating the code). Then the encoded signal is propagated towards a molecular receiver. We utilised mathematical models to describe how to create these binary codes and created several experiments in the Waterford Institute of Technology biology laboratory to show these same codes been produced by real bacterial cells.
Bacteria can exchange information among themselves through molecules and use it to act collectively. In this paper, we exploit this function, and use multiple bacterial populations to create binary codes (often used to correct errors in digital transmissions) to protect their emitted molecules from any unwanted environmental effect, that may reduce the amount of signal reaching the receiver.
Who will it help?
In a future when synthetic cells are applied to monitor and treat health-related issues in humans, animals and plants, the accuracy of the information collected and exchanged by them is fundamental. Molecular channel codes can ensure that synthetic cells do not add other challenges to these solutions, such as possible errors that could lead to incorrect diagnostics of a disease in the human gut. In addition to this, the same codes can help researchers to better understand the efficiency of the signalling processes that occur inside microbial communities which ensures their survival and might lead to development of chronic diseases in humans, animals and plants.
What is the future of this research?
The next step in this research, will be to improve our mathematical model to include the reception of the molecular encoded signals by other bacteria, electrochemical and photochemical systems. In addition, we will extend our experimental tests to investigate how the bacterial populations and signals would behave when mixed together from the beginning of the experiment. Finally, we will increase the number of distinct codes (currently, we only have four), which would make this molecular communications system more robust against channel-induced errors. We expect to utilise this model and results to support the development of realistic end-to-end molecular communications systems using bacteria, which have been investigated theoretically.
Despite this being “blue-sky” research, we envision the future commercialisation of engineered bacteria-based systems for Agri-Tech (to monitor cattle health and soil quality, for example) and medicine (specially to monitor and treat gut-related diseases). To this end, this technology needs to continuously evolve to be able to have a similar performance under a wide variety of channel conditions and effects. We also envision that the proposed molecular channel code scheme can inspire other researchers to develop novel molecular encoding techniques that can be applied to encrypt information collected and stored in bacteria’s plasmid. We believe this would be the fundamental step towards the development of biotechnology applications, which could revolutionise the pharmaceutical and Agri-tech industries.
Publication Title: Modulated Molecular Channel Coding Scheme for Multi-Bacterial Transmitters
Authors: Daniel Perez Martins, Jennifer Drohan, Sarah Foley, Lee Coffey and Sasitharan Balasubramaniam
Publication Date: 15 November 2021
Journal: SenSys ’21: Proceedings of the 19th ACM Conference on Embedded Networked Sensor Systems
Link to publication:




